|

Logistic flow of ArrayCore
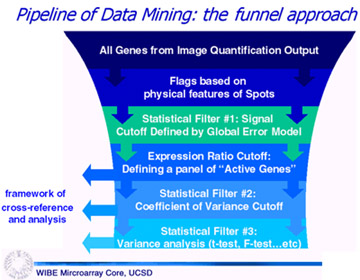
Data Processing Pipeline:
Data Management and Analysis Facilitaed by a centralized
mangagement system, users of the microarray facility
can acess and share (when permitted by the data owner)
their data through internet. Internet access to data
analysis software installed on our central server will
also be provided. Passwords are required to login
to the data management and analysis system. Guests are
welcomed to browse gene lists and public data via our
online webserver.
Logistically, Data from microarray experiments are
stored centrally on a database server for retrieval
and analysis though a web interface. You can download
data to your local system in Excel for local processing
and analysis, or process and analyze it directly on
the ArrayCore
server.
Data from slides processed through the ArrayCore facility
are automatically loaded into the server. Existing data
from a variety of array technologies, such as Affymetrix,
glass slides from other facilities, or nylon membrane
can be loaded. Because of the variations in data formats,
please contact us for assistance with loading your existing
data.
With data stored centrally, it can be reanalyzed as
new methods become available that may provide improved
sensitivity in detecting differentially expressed genes.
Collaborating laboratories can exchange and share data
from different experiments.
Our database server is based on the GeNet Expression
Data Management system, in complimentry of our main
data analysis interface, GeneSpring, both are developed
by Silicon
Genetics.
|


